

[HOME]
[WEB ALBUMS]
[PROJECTS]
[ARCHIVE]
[DOWNLOADS]
[LINKS]
project MK6 Adding more Python functions to Cfrad post processing
Introduction.
The extra functions are often just a few script lines, so it is not usefull to publish a new version of a plot script with every new line.
We just take the standard plot script here.
http://parac.eu/cfrad-plot-eng.zip
and indicate where to insert the new fuction. You have to find the equivalent location in your own script.
1. Average multiple spectra lines.
If you have a stable measurement, you could wish to integrate the signal of the source longer than 5 minutes.
Average of a number of 5 minute spectra and save sum file to disk for later use.
Insert after original line 189
#---------------------------------
sumsets=np.zeros((2048)) ;
sumf=3#sum spectrum number from
sumt=23#sum spectrum number to
for sets in range (sumf,sumt):
sumsets=sumsets+outsets[sets]
sumsets=sumsets/(sumt+1-sumf)
np.savetxt(filename+"sum"+str(sumf)+"-"+str(sumt)+".txt",sumsets)# save sum file
#---------------------------------
You can add the average spectrum sum at the bottom of the overview graph.
Insert after original line 192
#---------------------------------
ax.plot(f-Sf,sumsets,linewidth=1)#
#---------------------------------
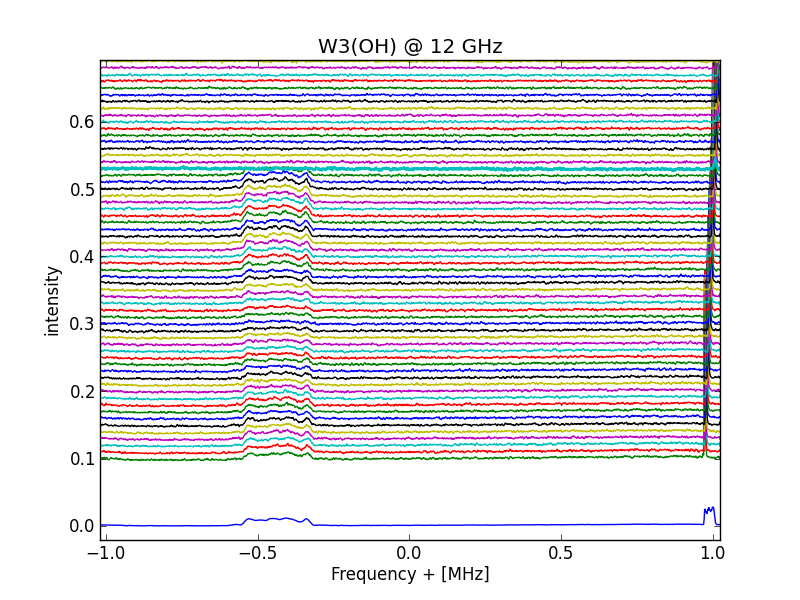
Adding multiple 5 minute spectra and plot result
2.Prepare for contour plotting or fits file generation.
To show a contour graph, all the nrad files have to be corrected for the bandpass curve.
Also the spectra base lines have to be normalized; that is; the baseline drift has to be compensated.
If not, you get jumps in the contour lines. See project MK5 to post process the contour graph.
Insert after original line 187
#-------------------------------
offsetcor=1-((data[50]+data[60]+data[1990]+data[2000])/4)#line space normalizing
data=data+offsetcor
np.savetxt(filename+str(sets)+".tct",data)#csv preparation. to disable; place # at start of line
#------------------------------
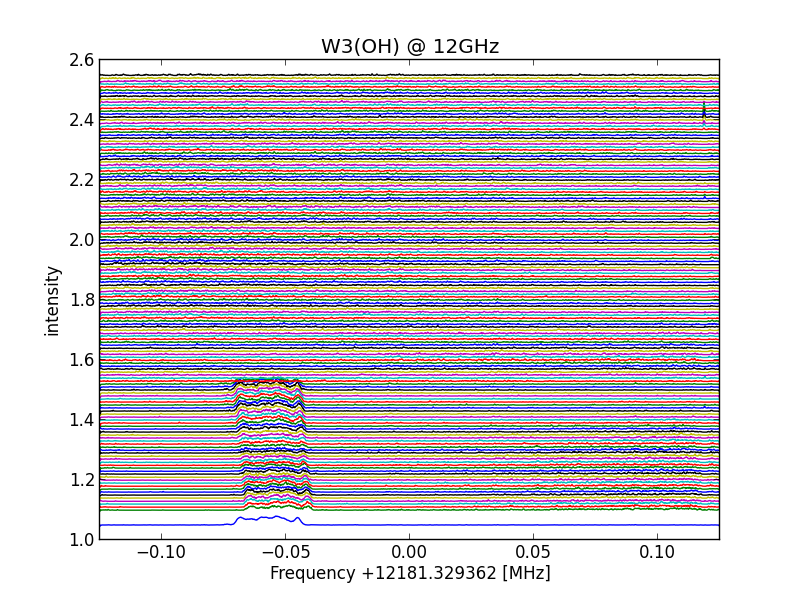
No Normalization
With normalization
-12GHz-slanted-corrected.png)
W3(OH-12GHz-slanted and corrected
4. Plot all spectra with 2048 BINS as x-axis.
This can be usefull when you want to know in which bin an amplitude effect took place.
Insert after original line 207
#------------------------------------------
fig=mpl.figure(2)
ax = fig.add_subplot(111)
#ax.plot(sumsets,linewidth=1) #average
for i in range (vans,ms+1):
ax.plot((outsets[i]+offsets[i]))
ax.set_title('signal')
ax.set_xlabel('BIN')
ax.set_ylabel('Intensity')
mpl.show()
#------------------------------------------
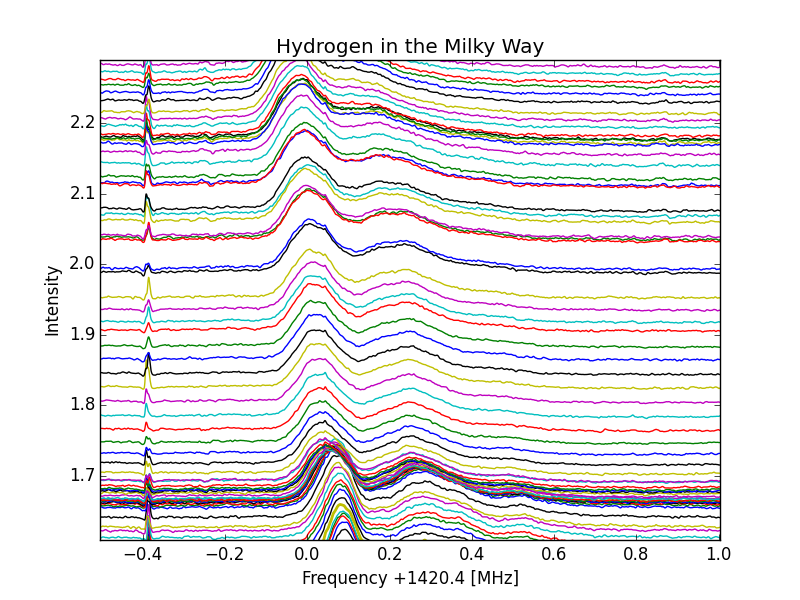
temperature frequency drifted lines

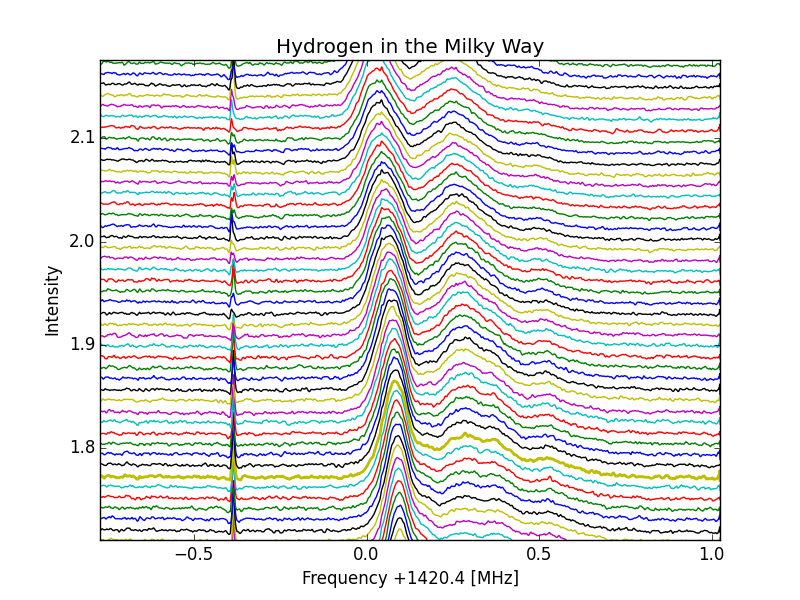
All lines shift corrected
6. Reverse spectrum.
There are several ways to do that, but I choose on obvious one.
Replace line 202 with
#------------------------------------------
ax.set_xlim( Sf, -Sf)
#------------------------------------------
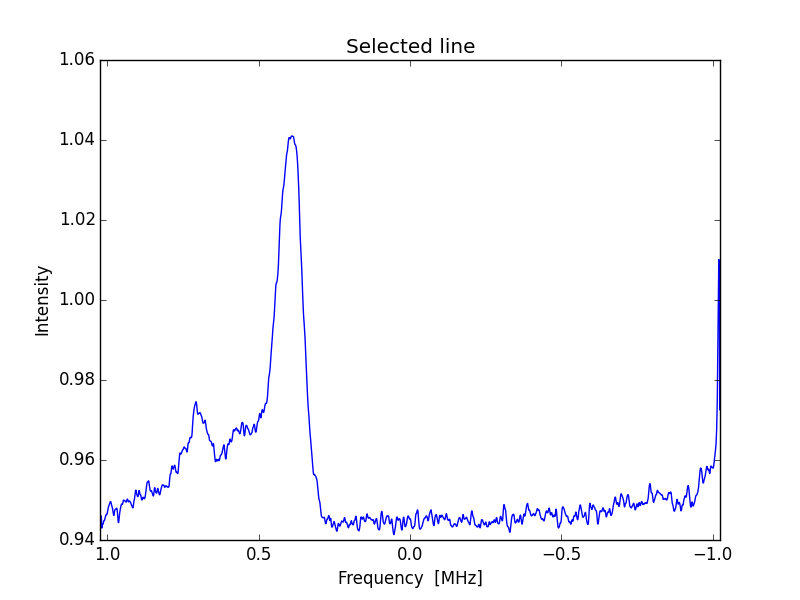
X scale corrected curve
7. Adding a second scale; for instance; VLSR.
For neutral hydrogen;
frVLSR=(3e8*(1-(obsfreq-bw/2)/restfreq)/1000-LSR
toVLSR=(3e8*(1-(obsfreq+bw/2)/restfreq)/1000-LSR
restfreq=1420 405 752 Hz
observation frequency =1420 000 000 Hz
bandwidth=2048000 Hz
LSR=Velocity of the Local Standard of Rest of the earth; this includes rearth own rotation and rotation around the sun.
This value can be calculated from;
http://neutronstar.joataman.net/technical/radial_vel_calc.html
This value varies between + and - 30 km/s; As a first appoximation; LSR can be set to 0.
Insert after original line 206
#------------------------------------------
ax2 = ax.twiny()
ax2.set_xlim( frVLSR, toVLSR)
ax2.set_xlabel('VLSR [km/s]')
ax2.set_title('Milky Way - 16-6-2014', y=1.07)
#------------------------------------------
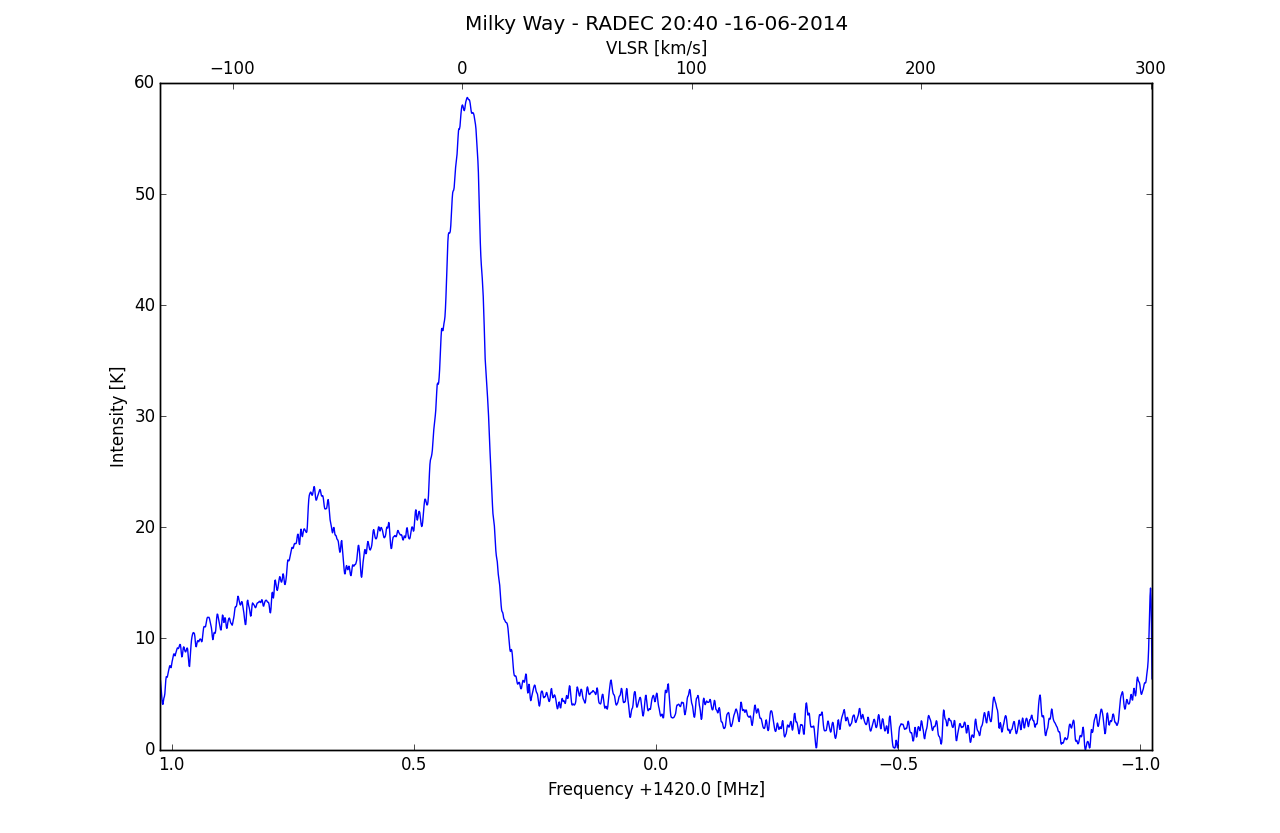
Added second X scale
8. Change the reference level from 1 to 0.
Replace line 141 with
#------------------------------------------
ax.plot(f-Sf,(raw4)-np.min(raw4), linewidth=1,label='clean')
#------------------------------------------
Or select a 'no signal' region and calculate an average reference value to subtract
#------------------------------------------
ax.plot(f-Sf,(raw4)-np.mean(raw4[300:400]), linewidth=1,label='clean')
#------------------------------------------
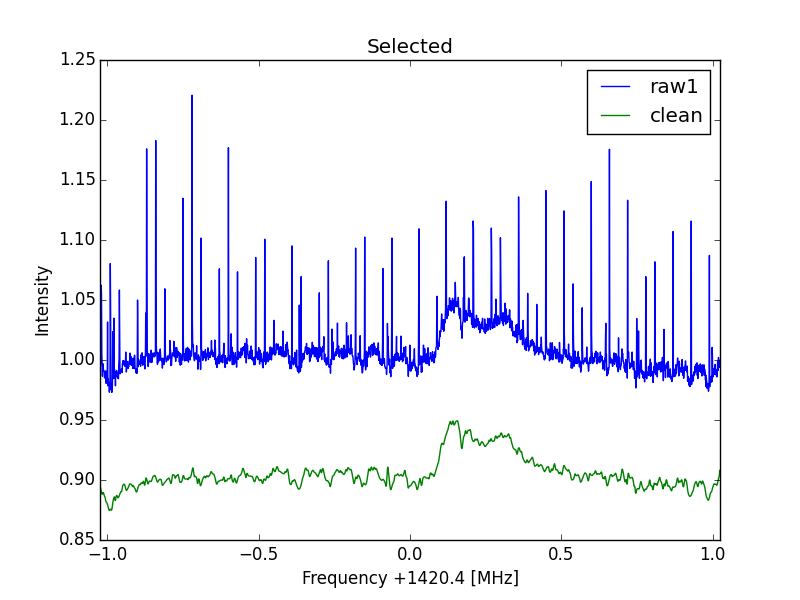
RFI suppression
10.Vertical Scale calibration.
Point telescope to strong H1 source like direction RA 20H, DEC=40deg.
Use https://www.astro.uni-bonn.de/hisurvey/profile/ and also fill in the beam of the telescope to get the profile.
Note the peak value of the unibonn curve =UB
Note your raw peak value=RV
Gain correction: GC=UB/RV
Insert after original line 150
#------------------------------------------
fig=mpl.figure(sets)
ax = fig.add_subplot(111)
ax.plot(f-Sf,GC*(raw4-np.min(raw4)), linewidth=1)
ax.set_xlim( -Sf, Sf)
ax.set_title('Selected line')
ax.set_xlabel('Frequency +1420.4 [MHz]') #
ax.set_ylabel('Brightness Temperature [K]')
mpl.show()
#------------------------------------------
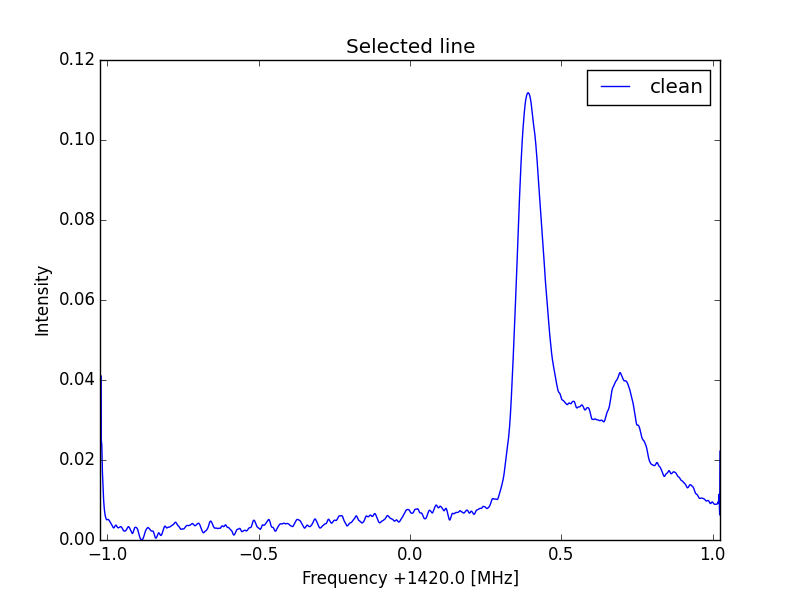
Standard curve
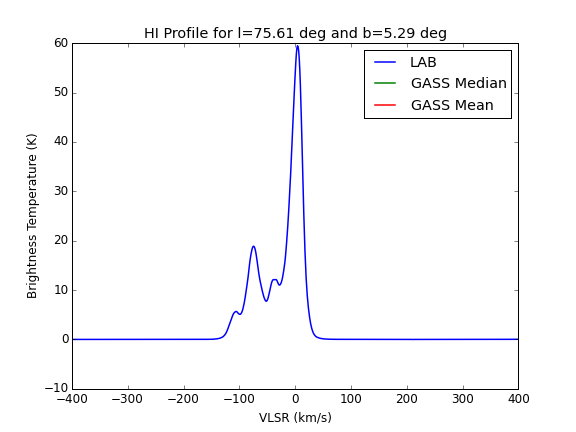
Reference curve
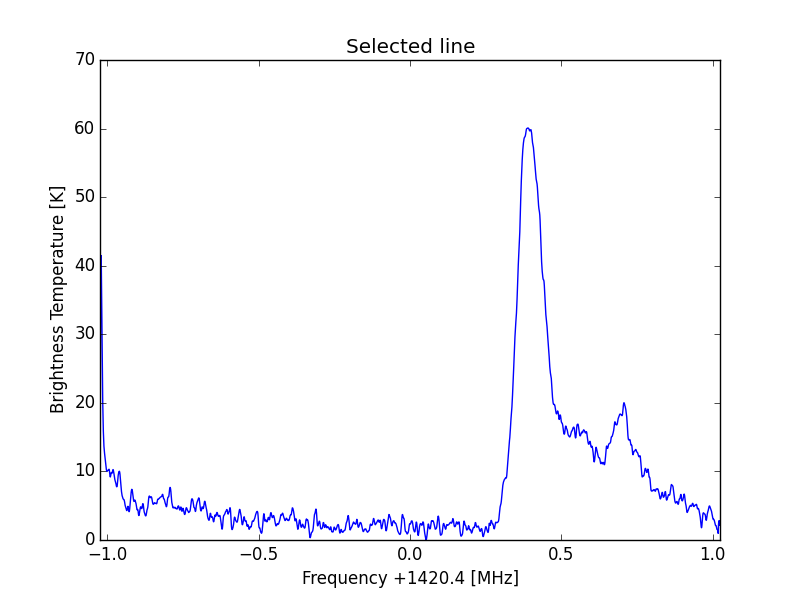
Y scale corrected curve
11. Intensity as a function of time.
The spectrum consists of 2048 bins; each bin can be selected as a time signal.
It will be more easy to choose a bin when option 4 is used.
More bins can be chosen also to form an averaging signal.
Insert after original line 207
#------------------------------------------
data=np.zeros((ms+1,2048)) ;
sign1=np.zeros((ms+1,1));#signal 1 of possible 2048 signals
reflow=np.zeros((ms+1,1));#reference
for n in range (0,ms+1):
data[n] = np.loadtxt(filename+str(n)+".txt")
data[n]=data[n]*bpcorr
for k in range (1407,1408):# select bins of interest
sign1[n]=sign1[n]+data[n,k]
for k in range (500,510): #select quiet part of spectrum (in bins)
reflow[n]=reflow[n]+data[n,k]
sign1=sign1/1
reflow=reflow/10
fig=mpl.figure(2)
ax = fig.add_subplot(111)
ax.plot(516*((sign1-reflow)-np.min(sign1-reflow) ) )#
ax.set_title('Milky Way drift scan')
ax.set_xlabel('*5min from start')
ax.set_ylabel('Brightness Temperature [K]')
mpl.show()
#------------------------------------------
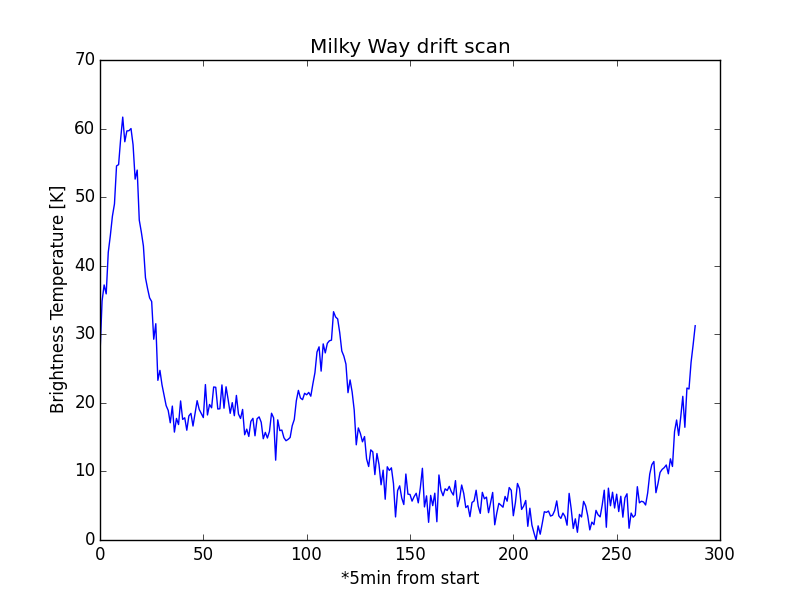
Brightness versus Time
Michiel Klaassen 03mrt2020